16. Generalized Linear Madness
Contents
16. Generalized Linear Madness¶
# Install packages that are not installed in colab
try:
import google.colab
%pip install -q watermark
%pip install git+https://github.com/ksachdeva/rethinking-tensorflow-probability.git
except:
pass
%load_ext watermark
# Core
import numpy as np
import arviz as az
import tensorflow as tf
import tensorflow_probability as tfp
# visualization
import matplotlib.pyplot as plt
from rethinking.data import RethinkingDataset
from rethinking.data import dataframe_to_tensors
from rethinking.mcmc import sample_posterior
# aliases
tfd = tfp.distributions
tfb = tfp.bijectors
Root = tfd.JointDistributionCoroutine.Root
2022-01-19 19:16:09.010782: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory
2022-01-19 19:16:09.010830: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
%watermark -p numpy,tensorflow,tensorflow_probability,arviz,scipy,pandas,rethinking
numpy : 1.21.5
tensorflow : 2.7.0
tensorflow_probability: 0.15.0
arviz : 0.11.4
scipy : 1.7.3
pandas : 1.3.5
rethinking : 0.1.0
# config of various plotting libraries
%config InlineBackend.figure_format = 'retina'
16.1 Geometric people¶
16.1.1 The scientific model¶
16.1.2 The statistical model¶
Code 16.1¶
Even though Generalized Linear Models are very powerful, not everything can still be modeled as a GLM - a linear combination of variables mapped onto a non-linear outcome.
Author strongly suggests that starting with GLM instead of real models may have severe consequences as it may be difficult to learn from failures in this case.
Here the focus is going to be using scientific information in modeling
d = RethinkingDataset.Howell1.get_dataset()
# scale observed variables
d["w"] = d.weight / d.weight.mean()
d["h"] = d.height / d.height.mean()
d
| height | weight | age | male | w | h | |
|---|---|---|---|---|---|---|
| 0 | 151.765 | 47.825606 | 63.0 | 1 | 1.343015 | 1.097650 |
| 1 | 139.700 | 36.485807 | 63.0 | 0 | 1.024577 | 1.010389 |
| 2 | 136.525 | 31.864838 | 65.0 | 0 | 0.894813 | 0.987425 |
| 3 | 156.845 | 53.041914 | 41.0 | 1 | 1.489497 | 1.134391 |
| 4 | 145.415 | 41.276872 | 51.0 | 0 | 1.159117 | 1.051723 |
| ... | ... | ... | ... | ... | ... | ... |
| 539 | 145.415 | 31.127751 | 17.0 | 1 | 0.874114 | 1.051723 |
| 540 | 162.560 | 52.163080 | 31.0 | 1 | 1.464818 | 1.175725 |
| 541 | 156.210 | 54.062497 | 21.0 | 0 | 1.518157 | 1.129798 |
| 542 | 71.120 | 8.051258 | 0.0 | 1 | 0.226092 | 0.514380 |
| 543 | 158.750 | 52.531624 | 68.0 | 1 | 1.475167 | 1.148169 |
544 rows × 6 columns
tdf = dataframe_to_tensors("Howell1", d, ["h", "w"])
2022-01-19 19:16:11.734030: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory
2022-01-19 19:16:11.734062: W tensorflow/stream_executor/cuda/cuda_driver.cc:269] failed call to cuInit: UNKNOWN ERROR (303)
2022-01-19 19:16:11.734085: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (fv-az272-145): /proc/driver/nvidia/version does not exist
2022-01-19 19:16:11.734400: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
Code 16.2¶
The first thing to notice about the parameters k and p is that they are multiplied in the model and the data have no way to estimate anything except their product
def model_16_1(h):
def _generator():
p = yield Root(tfd.Sample(tfd.Beta(2.0, 18.0, name="p"), sample_shape=1))
k = yield Root(tfd.Sample(tfd.Exponential(rate=0.5, name="k"), sample_shape=1))
sigma = yield Root(
tfd.Sample(tfd.Exponential(rate=1.0, name="sigma"), sample_shape=1)
)
term1 = p ** 2
term2 = h ** 3
mu = tf.math.log(3.141593 * k[..., tf.newaxis] * term1[..., tf.newaxis] * term2)
scale = sigma[..., tf.newaxis]
w = yield tfd.Independent(
tfd.LogNormal(loc=mu, scale=scale), reinterpreted_batch_ndims=1
)
return tfd.JointDistributionCoroutine(_generator, validate_args=False)
jdc_16_1 = model_16_1(tdf.h)
NUM_CHAINS_FOR_16_1 = 2
# Note - if I use zeros as the starting point
# then the sampled params are really bad
#
# So starting the init state sampled from the model
# itself
p_init, k_init, sigma_init, _ = jdc_16_1.sample()
init_state = [
tf.tile(p_init, (NUM_CHAINS_FOR_16_1,)),
tf.tile(k_init, (NUM_CHAINS_FOR_16_1,)),
tf.tile(sigma_init, (NUM_CHAINS_FOR_16_1,)),
]
bijectors = [
tfb.Identity(),
tfb.Exp(),
tfb.Exp(),
]
posterior_16_1, trace_16_1 = sample_posterior(
jdc_16_1,
observed_data=(tdf.w,),
params=["p", "k", "sigma"],
init_state=init_state,
bijectors=bijectors,
)
WARNING:tensorflow:@custom_gradient grad_fn has 'variables' in signature, but no ResourceVariables were used on the forward pass.
WARNING:tensorflow:@custom_gradient grad_fn has 'variables' in signature, but no ResourceVariables were used on the forward pass.
WARNING:tensorflow:@custom_gradient grad_fn has 'variables' in signature, but no ResourceVariables were used on the forward pass.
Code 16.3¶
h_seq = np.linspace(0, d.h.max(), num=30)
p_sample = trace_16_1.posterior["p"].values[0]
k_sample = trace_16_1.posterior["k"].values[0]
sigma_sample = trace_16_1.posterior["sigma"].values[0]
def compute_w_from_samples(h):
mu = tf.math.log(3.141593 * k_sample * p_sample ** 2 * h ** 3)
return tfd.LogNormal(loc=mu, scale=sigma_sample).sample().numpy()
w_sim = np.array(list(map(compute_w_from_samples, h_seq))).T
mu_mean = np.mean(w_sim, 0)
w_CI = np.percentile(w_sim, q=(5.5, 94.5), axis=0)
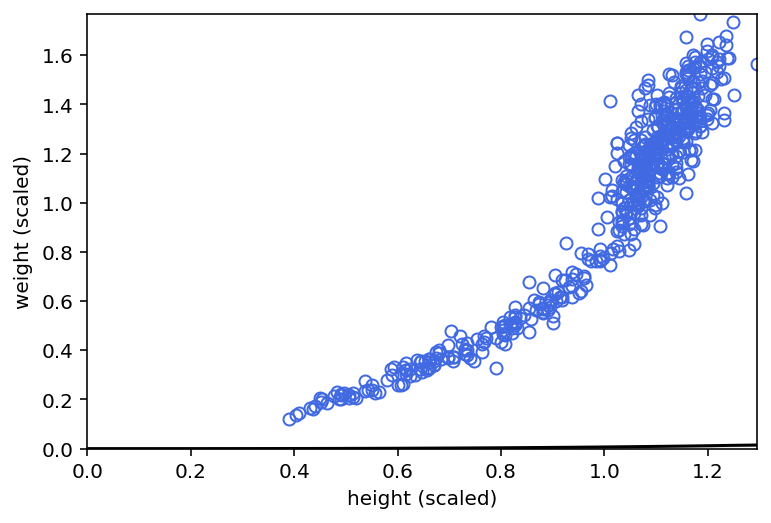
plt.plot(d.h, d.w, "o", c="royalblue", mfc="none")
plt.gca().set(
xlim=(0, d.h.max()),
ylim=(0, d.w.max()),
xlabel="height (scaled)",
ylabel="weight (scaled)",
)
plt.plot(h_seq, mu_mean, "k")
plt.fill_between(h_seq, w_CI[0], w_CI[1], color="k", alpha=0.2)
<matplotlib.collections.PolyCollection at 0x7fde34415d90>
Code 16.7 (Not working !)¶
Having issues with shape again :(
d.describe()
| y | gender | age | majority_first | culture | |
|---|---|---|---|---|---|
| count | 629.000000 | 629.000000 | 629.000000 | 629.000000 | 629.000000 |
| mean | 2.120827 | 1.505564 | 8.030207 | 0.484897 | 3.751987 |
| std | 0.727986 | 0.500367 | 2.497906 | 0.500170 | 1.960319 |
| min | 1.000000 | 1.000000 | 4.000000 | 0.000000 | 1.000000 |
| 25% | 2.000000 | 1.000000 | 6.000000 | 0.000000 | 3.000000 |
| 50% | 2.000000 | 2.000000 | 8.000000 | 0.000000 | 3.000000 |
| 75% | 3.000000 | 2.000000 | 10.000000 | 1.000000 | 5.000000 |
| max | 3.000000 | 2.000000 | 14.000000 | 1.000000 | 8.000000 |
def make_boxes_model(N, y, majority_first):
def _generator():
# prior
p = yield Root(
tfd.Sample(
tfd.Dirichlet(tf.cast(np.repeat(4, 5), dtype=tf.float32)),
sample_shape=1,
)
)
# probability of data
phi = [None] * 5
phi[0] = np.where(y == 2, 1, 0) # majority
phi[1] = np.where(y == 3, 1, 0) # minority
phi[2] = np.where(y == 1, 1, 0) # maverick
phi[3] = np.repeat(1.0 / 3.0, N) # random
phi[4] = np.where(
majority_first == 1, # follow first
np.where(y == 2, 1, 0),
np.where(y == 3, 1, 0),
)
# ps = tf.squeeze(p)
ps = p
phi = np.array(phi, dtype=np.float32)
print(ps.shape)
print(phi.shape)
# compute log( p_s * Pr(y_i|s )
for j in range(5):
phi[j] = tf.math.log(ps[j]) + tf.math.log(phi[j])
# compute average log-probability of y
logprob = tf.math.reduce_logsumexp(tf.stack(tf.constant(phi), axis=1), axis=1)
w = yield tfd.Independent(
tfd.Deterministic(loc=logprob), reinterpreted_batch_ndims=1
)
return tfd.JointDistributionCoroutine(_generator, validate_args=False)
# dat= dict(N=d.shape[0],
# y=d.y.values,
# majority_first=d.majority_first.values)
tdf = dataframe_to_tensors("Boxes", d, ["y", "majority_first"])
jdc_boxes_model = make_boxes_model(d.shape[0], tdf.y, tdf.majority_first)
# uncomment the line below to see the error
# jdc_boxes_model.sample(2)
Code 16.8 (depends on 16.7) [Not implemented]¶
16.3 Ordinary differential nut cracking¶
Code 16.9¶
Modelling nut opening skill of chimpanzees
d = RethinkingDataset.PandaNuts.get_dataset()
d
| chimpanzee | age | sex | hammer | nuts_opened | seconds | help | |
|---|---|---|---|---|---|---|---|
| 0 | 11 | 3 | m | G | 0 | 61.0 | N |
| 1 | 11 | 3 | m | G | 0 | 37.0 | N |
| 2 | 18 | 4 | f | wood | 0 | 20.0 | N |
| 3 | 18 | 4 | f | G | 0 | 14.0 | y |
| 4 | 18 | 4 | f | L | 0 | 13.0 | N |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 79 | 10 | 15 | m | G | 7 | 12.0 | N |
| 80 | 10 | 15 | m | G | 5 | 12.5 | N |
| 81 | 6 | 15 | m | G | 8 | 13.0 | N |
| 82 | 6 | 16 | m | G | 24 | 20.0 | N |
| 83 | 6 | 16 | m | G | 25 | 36.0 | N |
84 rows × 7 columns
We want to understand how nut opening skills develop and which factors contribute to it.
We need a generative model of nut opening rate as it varies by age.
On simple factor to assume is the individual’s strenght and as age increases it increases as well.
Code 16.10¶
N = int(1e4)
phi = tfd.LogNormal(np.log(1), 0.1).sample((N,)).numpy()
k = tfd.LogNormal(np.log(2), 0.25).sample((N,)).numpy()
theta = tfd.LogNormal(np.log(5), 0.25).sample((N,)).numpy()
# relative grow curve
ax = plt.subplot(xlim=(0, 1.5), ylim=(0, 1), xlabel="age", ylabel="body mass")
at = np.array([0, 0.25, 0.5, 0.75, 1, 1.25, 1.5])
ax.set(xticks=at, xticklabels=np.round(at * d.age.max()))
x = np.linspace(0, 1.5, 101)
for i in range(20):
plt.plot(x, 1 - np.exp(-k[i] * x), "gray", lw=1.5)
plt.show()
# implied rate of nut opening curve
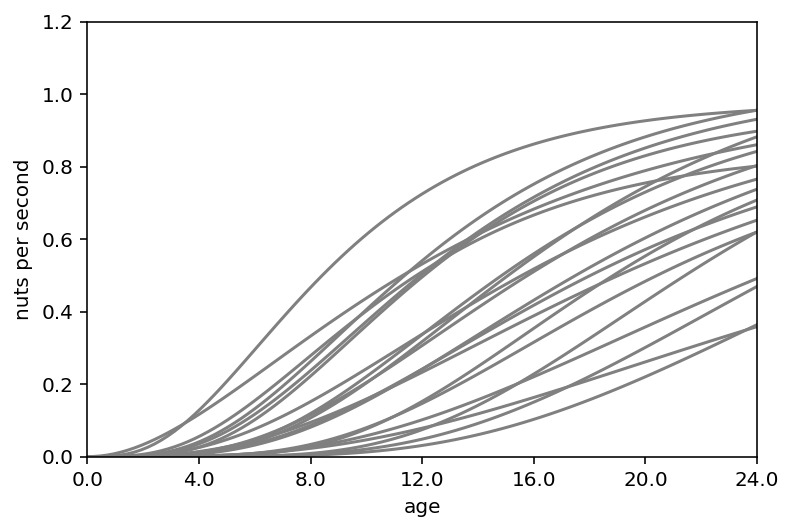
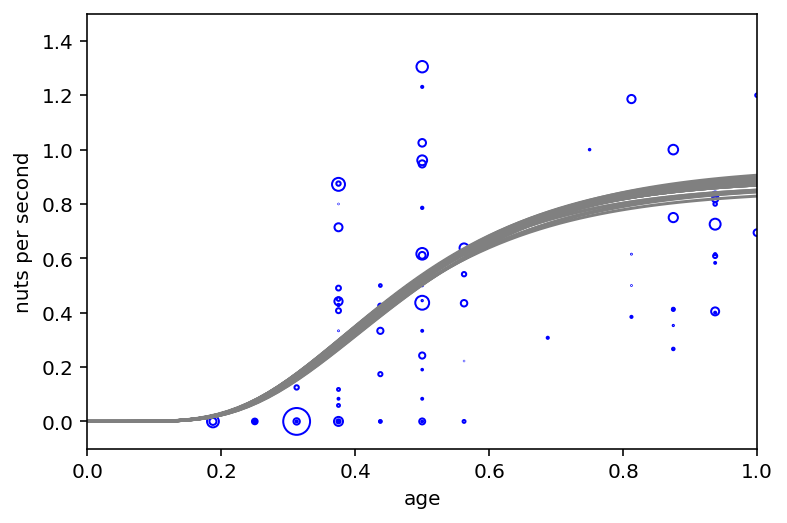
ax = plt.subplot(xlim=(0, 1.5), ylim=(0, 1.2), xlabel="age", ylabel="nuts per second")
at = np.array([0, 0.25, 0.5, 0.75, 1, 1.25, 1.5])
ax.set(xticks=at, xticklabels=np.round(at * d.age.max()))
x = np.linspace(0, 1.5, 101)
for i in range(20):
plt.plot(x, phi[i] * (1 - np.exp(-k[i] * x)) ** theta[i], "gray", lw=1.5)
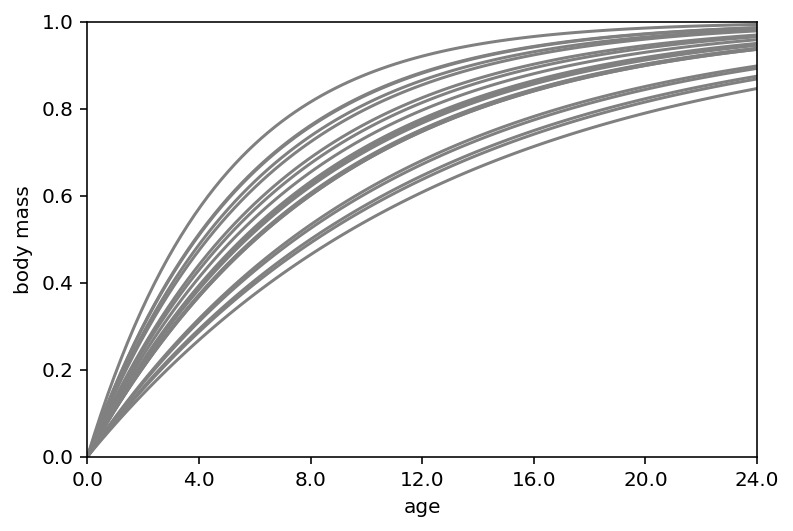
The first graph (body mass vs age) is showing that opening kill tries to start leveling off around age 12
The second graph (nuts per second vs age) shows many different developmental patterns
Code 16.11¶
d["age_prop"] = d.age.values / d.age.max()
tdf = dataframe_to_tensors("Nuts", d, ["nuts_opened", "age_prop", "seconds"])
def model_16_4(seconds, age):
def _generator():
phi = yield Root(
tfd.Sample(tfd.LogNormal(tf.math.log(1.0), 0.1, name="phi"), sample_shape=1)
)
k = yield Root(
tfd.Sample(tfd.LogNormal(tf.math.log(2.0), 0.25, name="k"), sample_shape=1)
)
theta = yield Root(
tfd.Sample(
tfd.LogNormal(tf.math.log(5.0), 0.25, name="theta"), sample_shape=1
)
)
term1 = phi[..., tf.newaxis]
term2 = (1 - tf.exp(-k[..., tf.newaxis] * age)) ** theta[..., tf.newaxis]
lambda_ = seconds * term1 * term2
n = yield tfd.Independent(tfd.Poisson(lambda_), reinterpreted_batch_ndims=1)
return tfd.JointDistributionCoroutine(_generator, validate_args=False)
jdc_16_4 = model_16_4(tdf.seconds, tdf.age_prop)
jdc_16_4.sample(2)
WARNING:tensorflow:From /opt/hostedtoolcache/Python/3.7.12/x64/lib/python3.7/site-packages/tensorflow_probability/python/distributions/distribution.py:342: calling Poisson.__init__ (from tensorflow_probability.python.distributions.poisson) with interpolate_nondiscrete is deprecated and will be removed after 2021-02-01.
Instructions for updating:
The `interpolate_nondiscrete` flag is deprecated; instead use `force_probs_to_zero_outside_support` (with the opposite sense).
StructTuple(
var0=<tf.Tensor: shape=(2, 1), dtype=float32, numpy=
array([[0.8813147],
[0.9588341]], dtype=float32)>,
var1=<tf.Tensor: shape=(2, 1), dtype=float32, numpy=
array([[2.0577004],
[2.9271986]], dtype=float32)>,
var2=<tf.Tensor: shape=(2, 1), dtype=float32, numpy=
array([[3.5044107],
[5.081681 ]], dtype=float32)>,
var3=<tf.Tensor: shape=(2, 1, 84), dtype=float32, numpy=
array([[[ 4., 3., 2., 0., 0., 1., 0., 7., 3., 1., 1., 7.,
0., 3., 3., 1., 4., 6., 1., 1., 2., 1., 1., 1.,
3., 0., 2., 2., 2., 4., 1., 0., 0., 4., 7., 9.,
3., 5., 4., 0., 1., 3., 4., 0., 4., 9., 7., 6.,
9., 16., 17., 0., 1., 2., 5., 0., 4., 12., 7., 11.,
0., 7., 1., 6., 20., 2., 2., 17., 23., 6., 2., 7.,
8., 28., 20., 17., 17., 10., 5., 5., 10., 8., 14., 15.]],
[[ 0., 0., 0., 0., 0., 2., 0., 10., 3., 2., 1., 10.,
1., 1., 2., 0., 3., 3., 3., 2., 1., 0., 1., 2.,
3., 1., 1., 3., 2., 3., 5., 2., 1., 3., 13., 7.,
0., 1., 3., 2., 1., 7., 11., 2., 2., 17., 12., 17.,
10., 20., 16., 1., 2., 9., 2., 7., 7., 18., 6., 14.,
1., 8., 4., 10., 28., 3., 1., 37., 32., 15., 5., 10.,
5., 39., 19., 19., 29., 25., 8., 6., 11., 12., 17., 28.]]],
dtype=float32)>
)
NUM_CHAINS_FOR_16_4 = 2
# Note - if I use zeros as the starting point
# then the sampled params are really bad
#
# So starting the init state sampled from the model
# itself
phi_init, k_init, theta_init, _ = jdc_16_4.sample()
init_state = [
tf.tile(phi_init, (NUM_CHAINS_FOR_16_4,)),
tf.tile(k_init, (NUM_CHAINS_FOR_16_4,)),
tf.tile(theta_init, (NUM_CHAINS_FOR_16_4,)),
]
bijectors = [
tfb.Identity(),
tfb.Identity(),
tfb.Identity(),
]
posterior_16_4, trace_16_4 = sample_posterior(
jdc_16_4,
observed_data=(tdf.nuts_opened,),
params=["phi", "k", "theta"],
init_state=init_state,
bijectors=bijectors,
)
Code 16.12¶
post_phi = trace_16_4.posterior["phi"].values[0]
post_k = trace_16_4.posterior["k"].values[0]
post_theta = trace_16_4.posterior["theta"].values[0]
plt.subplot(xlim=(0, 1), ylim=(-0.1, 1.5), xlabel="age", ylabel="nuts per second")
at = np.array([0, 0.25, 0.5, 0.75, 1, 1.25, 1.5])
ax.set(xticks=at, xticklabels=np.round(at * d.age.max()))
# raw data
pts = d.nuts_opened.values / d.seconds.values
normalize = lambda x: (x - np.min(x)) / (np.max(x) - np.min(x))
point_size = normalize(d.seconds.values)
eps = (d.age_prop[1:] - d.age_prop[:-1]).max() / 5
jitter = eps * tfd.Uniform(low=-1, high=1).sample(d.age_prop.shape)
plt.scatter(
d.age_prop.values + jitter,
pts,
s=(point_size * 3) ** 2 * 20,
color="b",
facecolors="none",
)
# 30 posterior curves
x = np.linspace(0, 1.5, 101)
for i in range(30):
plt.plot(x, post_phi[i] * (1 - np.exp(-post_k[i] * x)) ** post_theta[i], "gray")
16.4 Population dynamics¶
Code 16.13¶
d = RethinkingDataset.LynxHare.get_dataset()
plt.plot(np.arange(1, 22), d.iloc[:, 2], "ko-", lw=1.5)
plt.gca().set(ylim=(0, 90), xlabel="year", ylabel="throusands of pelts")
at = np.array([1, 11, 21])
plt.gca().set(xticks=at, xticklabels=d.Year.iloc[at - 1])
plt.plot(np.arange(1, 22), d.iloc[:, 1], "bo-", lw=1.5)
plt.annotate("Lepus", (17, 80), color="k", ha="right", va="center")
plt.annotate("Lynx", (19, 50), color="b", ha="right", va="center")
plt.show()
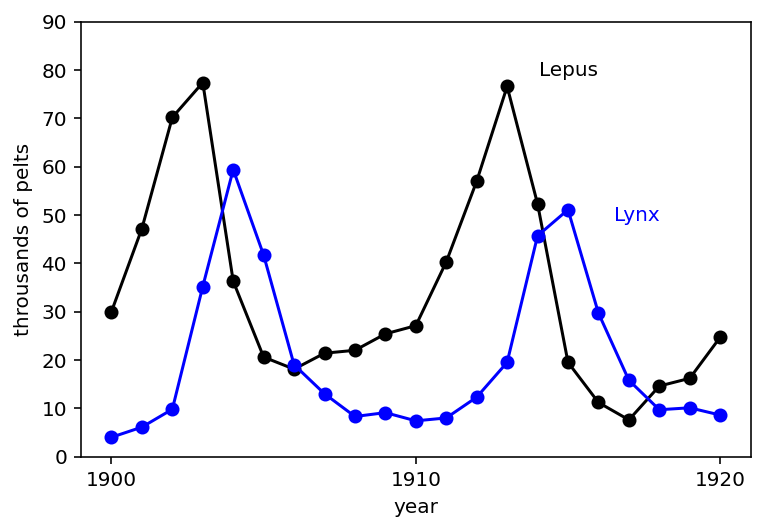
Above graph is showing 20 years of Lynx & hare pelts (attacks).
They seem to be related to each other. Both fluctuate but they seem to fluctuate together
Code 16.14¶
def sim_lynx_hare(n_steps, init, theta, dt=0.002):
L0 = init[0]
H0 = init[1]
def f(val, i):
H, L = val
H_i = H + dt * H * (theta[0] - theta[1] * L)
L_i = L + dt * L * (theta[2] * H - theta[3])
return (H_i, L_i)
(H, L) = tf.scan(f, np.arange(n_steps - 1), (H0, L0))
H = np.concatenate([np.expand_dims(H0, -1), H.numpy()])
L = np.concatenate([np.expand_dims(L0, -1), L.numpy()])
return np.stack([L, H], axis=1)
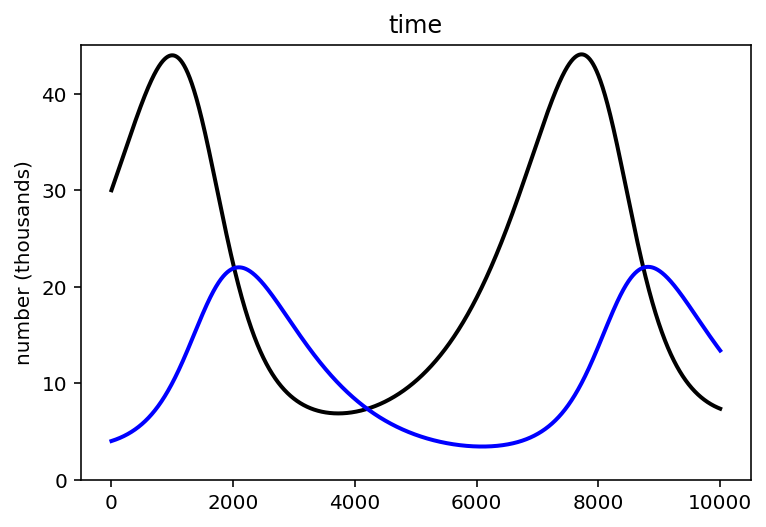
Code 16.15¶
theta = np.array([0.5, 0.05, 0.025, 0.5])
z = sim_lynx_hare(int(1e4), d.values[0, 1:3], theta)
plt.plot(z[:, 1], color="black", lw=2) # Hare population
plt.plot(z[:, 0], color="blue", lw=2) # Lynx population
plt.gca().set(ylim=(0, np.max(z[:, 1]) + 1), ylabel="number (thousands)", xlabel="")
plt.title("time")
plt.show()
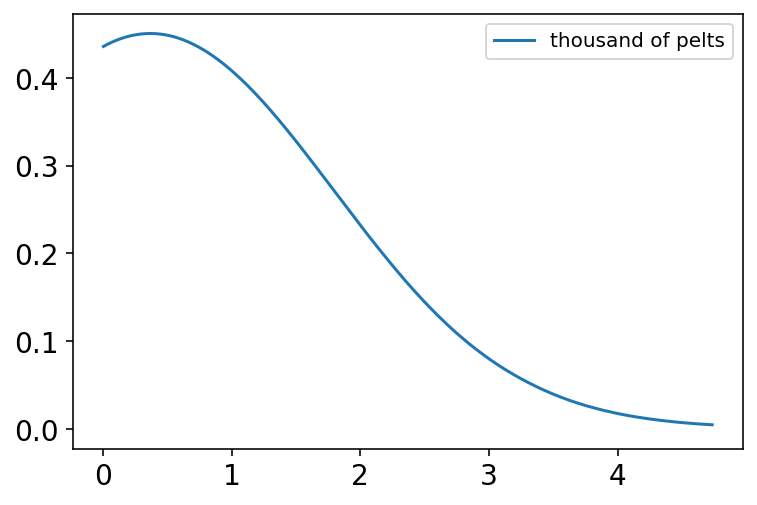
Code 16.16¶
N = int(1e4)
Ht = int(1e4)
p = tfd.Beta(2, 18).sample((N,)).numpy()
h = tfd.Binomial(Ht, probs=p).sample().numpy()
h = np.round(h / 1000, 2)
az.plot_kde(h, label="thousand of pelts", bw=1)
plt.show()
Code 16.17¶
Ordinary differential equations